Research

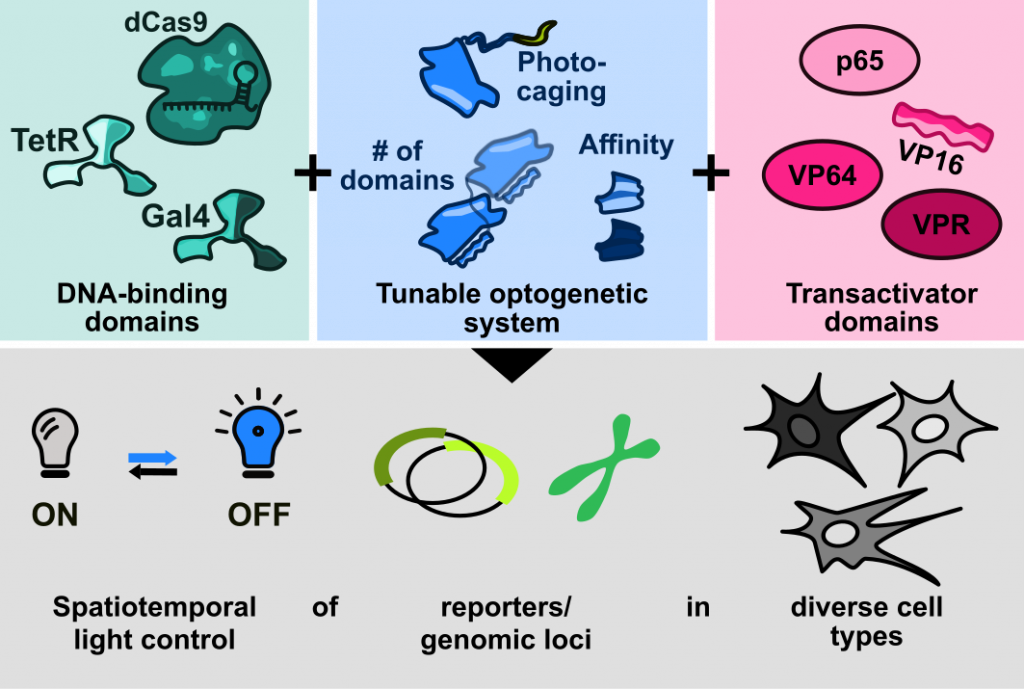
The goal of our group is to develop methods for the control of proteins by various exogenous stimuli. We want to address one of the main challenges Synthetic Biology and Biotechnology are facing, the persisting lack of precise methods for the flexible and reversible control of protein activity in living cells. To this end, we focus on the exploration of physical stimuli beyond light that will open up new avenues for the non-invasive regulation of molecular pathways.
Our research builds on a variety of methods, centered around the combination of rational protein engineering with high-throughput screening methods to engineer and optimize proteins.
TEAM

Dr. Jan Mathony
Junior Group Leader
Research Interest: Switchable Proteins, Allostery, CRISPR
Hobbies: Hiking, Photography, Music
Download CV

Ann-Sophie Kröll
Ph.D. Student
Research Interest: Genome Editing, Protein Engineering, Protein Switches
Hobbies: Quidditch, Basketball

Pegi Shehu
Master Student, Biomolecular Engineering
Research Interest: Protein Design & Engineering, Antibodies, Immunology
Hobbies: Reading, Singing, Coffee time, Traveling

Joshua Eigenmann
Master Student, MobiTech

Nele Pascale Happ
Bachelor Student, MobiTech
Research Interest: Genome Editing, Thermal Protein Switches
Hobbies: Swimming, Hiking, Books

Kira Hoffmann
Bachelor Student, MoBiTech
Research Interest: Protein Engineering, Thermal Protein Switches
Hobbies: Reading, Volleyball, Puzzles
Publications
Journal Papers
Muench, P., Fiumara, M., Southern, N., Coda, D., Aschenbrenner, S., Correia, B., Graeff, J., Niopek, D., and Mathony, J. (2023). A modular toolbox for the optogenetic deactivation of transcription. bioRxiv. https://doi.org/10.1101/2023.11.06.565805.
Mathony, J., and Niopek, D. (2023). Domäneninsertion: Untersuchung zur Konstruktion schaltbarer Hybridproteine. BIOspektrum 29, 595–598. https://doi.org/10.1007/s12268-023-2032-6.
Mathony, J., Aschenbrenner, S., Becker, P., and Niopek, D. (2023). Dissecting the Determinants of Domain Insertion Tolerance and Allostery in Proteins. Advanced science (Weinheim, Baden-Wurttemberg, Germany) 10, e2303496. https://doi.org/10.1002/advs.202303496.
Adam, L., Stanifer, M., Springer, F., Mathony, J., Brune, M., Di Ponzio, C., Eils, R., Boulant, S., Niopek, D., and Kallenberger, S.M. (2023). Transcriptomics-inferred dynamics of SARS-CoV-2 interactions with host epithelial cells. Science signaling 16. https://doi.org/10.1126/scisignal.abl8266.
Stadelmann, T., Heid, D., Jendrusch, M., Mathony, J., Rosset, S., Correia, B.E., and Niopek, D. (2021). A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins. bioRxiv, 2021‐2008. https://doi.org/10.1101/2021.08.21.457204.
Mathony, J., and Niopek, D. (2021). Enlightening allostery: designing switchable proteins by photoreceptor fusion. Advanced Biology 5, 2000181. https://doi.org/10.1002/adbi.202000181.
Hoffmann, M.D., Mathony, J., Upmeier zu Belzen, J., Harteveld, Z., Aschenbrenner, S., Stengl, C., Grimm, D., Correia, B.E., Eils, R., and Niopek, D. (2021). Optogenetic control of Neisseria meningitidis Cas9 genome editing using an engineered, light-switchable anti-CRISPR protein. Nucleic acids research 49, e29. https://doi.org/10.1093/nar/gkaa1198.
Adam, L., Stanifer, M., Springer, F., Mathony, J., Di Ponzio, C., Eils, R., Boulant, S., Niopek, D., and Kallenberger, S.M. (2021). Dynamics of SARS-CoV-2 host cell interactions inferred from transcriptome analyses. https://doi.org/10.1101/2021.07.04.450986.
Mathony, J., Hoffmann, M.D., and Niopek, D. (2020). Optogenetics and CRISPR: a new relationship built to last. Photoswitching Proteins: Methods and Protocols, 261–281. https://doi.org/10.1007/978-1-0716-0755-8_18.
Mathony, J., Harteveld, Z., Schmelas, C., Upmeier zu Belzen, J., Aschenbrenner, S., Sun, W., Hoffmann, M.D., Stengl, C., Scheck, A., Georgeon, S., et al. (2020). Computational design of anti-CRISPR proteins with improved inhibition potency. Nature chemical biology 16, 725–730. https://doi.org/10.1038/s41589-020-0518-9.
Upmeier zu Belzen, J., Bürgel, T., Holderbach, S., Bubeck, F., Adam, L., Gandor, C., Klein, M., Mathony, J., Pfuderer, P., Platz, L., et al. (2019). Leveraging implicit knowledge in neural networks for functional dissection and engineering of proteins. Nature Machine Intelligence 1, 225–235. https://doi.org/10.1038/s42256-019-0049-9.
Bubeck, F., Hoffmann, M.D., Harteveld, Z., Aschenbrenner, S., Bietz, A., Notbohm, J., Stengl, C., Mathony, J., Waldhauer, M.C., Dietz, L., et al. (2019). Engineered Anti-CRISPR Proteins for Precision Control of CRISPR-Cas9. Molecular Therapy 27, 297.
Contact
Jan Mathony
Institute of Pharmacy and Molecular Biotechnology (IPMB)
Heidelberg University
Im Neuenheimer Feld 364
4th floor, room 408
69120 Heidelberg, Germany
Email: jan.mathony@uni-Heidelberg.de
